Spatiotemporal FBA#
The incorporation of genome-scale metabolic reconstructions within spatiotemporal models that account for both spatial and temporal variations in the environment is desirable to connect genes to metabolic phenotype and system function [Chen et al., 2016].
COMETS and COMETSpy#
COMETS is a software platform for performing computer simulations of spatially structured microbial communities. It is based on stoichiometric modeling of the genome-scale metabolic network of individual microbial species using dynamic flux balance analysis, and on a discrete approximation of diffusion. COMETS is built and maintained by the Daniel Segre Lab at Boston University [Harcombe et al., 2014].
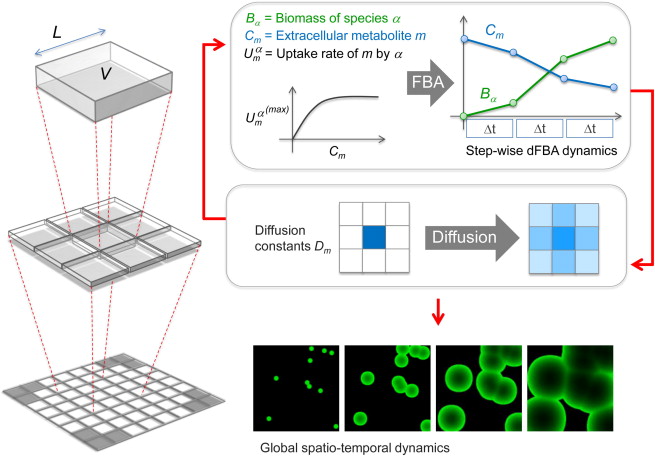
Fig. 7 A Schematic Representation of the Key Steps of COMETS Simulations.#
Since the MATLAB toolbox for COMETS is no longer actively maintained, we recommend installing its Python counterpart, COMETSPy, which serves as the Python interface for running COMETS simulations. Similar to the MATLAB interface for Gurobi, COMETSPy is not a standalone package. It is intended to be used alongside COMETS, which must be installed on the user’s computer beforehand. Additionally, Gurobi is required as a dependency for running COMETS simulations.

